Project 2.1
- PhD student: Miriam Mulorz
- Supervisor: Julian König
- Co-Supervisors: Joachim Burger, Thomas Hankeln
- Further TAC-members: René Ketting , Michaela Müller-McNicoll
- Research Group
My vision is to understand the impact of novel candidate splicing regulators, how they influence splicing decision and kinetics – and how these processes are is conserved across species.
As a post-transcriptional regulation mechanism, splicing is a keystone of gene expression. During splicing, the introns, long non-coding parts of the pre-mRNA, are excised, leaving only the exons behind, which make up the mature mRNA. Splicing is vital for proper development and tissue identity but also harbors the potential for evolutionary and proteomic diversity. The complexity of splicing calls for a carefully balanced regulation, which is facilitated by a multitude of RNA-binding proteins (RBPs) and small nuclear ribonucleoproteins (snRNPs) which together, build the spliceosome. The components of the spliceosome act on cis-regulatory sequences within the mRNA to facilitate and regulate splicing.
Although splicing is a highly conserved process in all eukaryotic organisms, it differs significantly from species to species. For example, while the yeast RNA contains only a few, rather short introns, mammals express long introns that separate short exons. In addition, the strength of splice sites and other cis-regulatory signals is decreases from lower to higher eukaryotes, making room for more complex regulation by trans-acting RBPs.
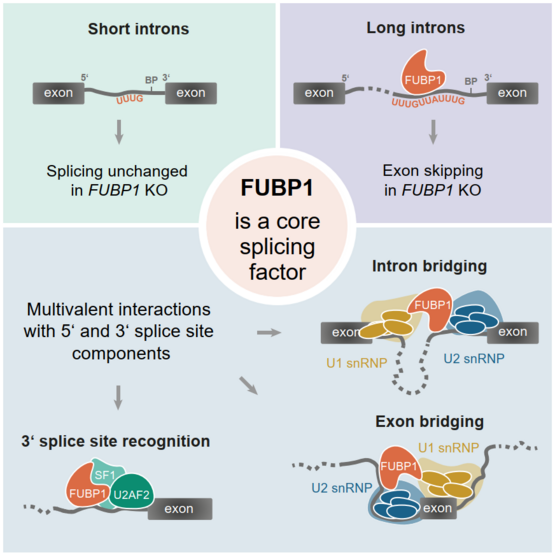
Our lab recently found that FUBP1, a well-known transcription factor and RNA-binding protein, is a new trans-acting RBP that modulates the binding of other splicing factors. My aim is to characterize the role of FUBP1 in splicing. To this end, I use high-throughput approaches to understand how FUBP1 affects the function of adjacent proteins as well as splicing decisions and kinetics. Moreover, I strive to unravel the evolutionary emergence of FUBP1. In particular, I focus on how the differences in splicing across species affect the conservation of FUBP1.