Project 2.2
- PhD student: Sarah Hallstein
- Supervisor: Julian König
- Co-Supervisors: Claudia Keller Valsecchi
- Further TAC-members: René Ketting
- Research Group
RNA modifications play an important role in gene regulation. N6-methyladenosine, the most abundant mRNA modification, is a key factor that influences mRNA fate and function. However, many aspects of this modification are unknown und not fully clear. I aim to provide a better understanding of m6A and a deeper insight into the underlying mechanisms mediated by this modification in mammalian evolution.
Similar to DNA, RNA can be chemically modified. These naturally occurring RNA modifications have been shown to play a crucial role in gene regulation, leading to a rapidly expanding research field known as epitranscriptomics. RNA modifications are essential for many cellular processes and can affect RNA function and fate. Until today, a variety of different RNA modifications has been discovered; however, not all of them are fully understood.
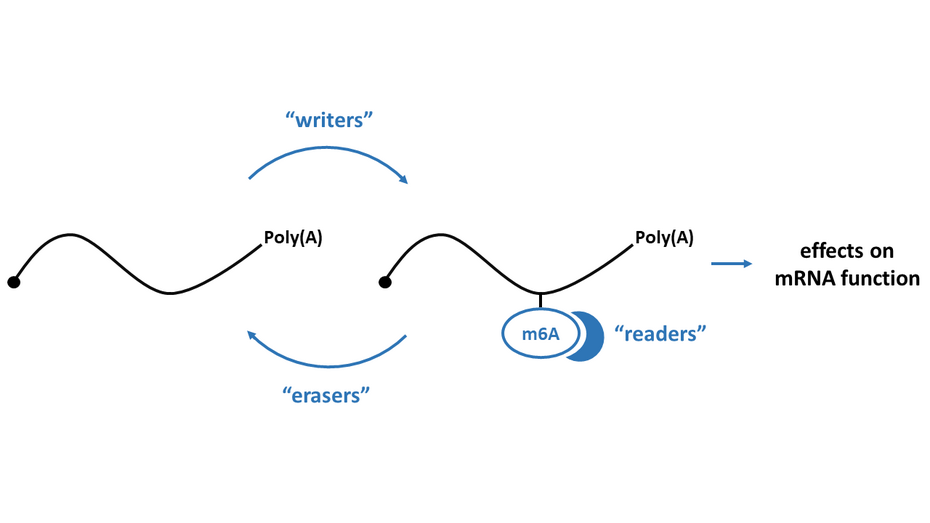
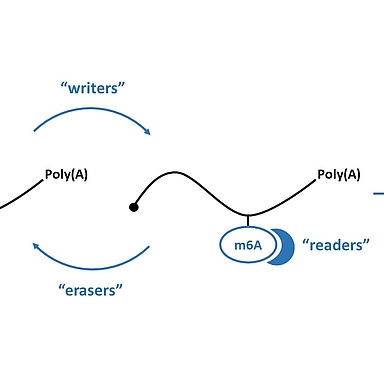
N6-methyladenosine (m6A) is the most abundant mRNA modification, which is predominantly deposited co-transcriptionally in the nucleus by the METTL3 writer complex. This epitranscriptomic mark is kept in the cytoplasm and various “reader” proteins in the nucleus or cytoplasm can bind to m6A. To better understand the importance and function of m6A, various mapping techniques have been developed. In our lab, we established the miCLIP2 protocol to map m6A in a transcriptome-wide manner using next generation sequencing and machine learning. Importantly, these mapping approaches have revealed m6A as a major contributor to RNA metabolism, influencing several molecular processes such as mRNA stability, splicing, translation and nuclear export. In this context, mediating mRNA instability is the most prominent consequence. Additionally, m6A seems to be of importance for other biological processes such as cellular differentiation and cancer. Nevertheless, many aspects of m6A on mRNA biology and the underlying mechanisms promoted by this modification are still unclear.
My aim is to provide a better understanding of m6A in post-transcriptional gene regulation and evolution with a particular interest in mammals. To that end, we want to focus on the deposition of m6A on mRNA, its functions and the underlying molecular mechanisms leading to the observed functions. Therefore, I will combine molecular biology techniques, computational approaches and functional genomics such as high-throughput sequencing. Using these different approaches allows us to place our findings into a broader context and gain a larger picture of the role of m6A.