Project 6.1
- PhD student: Alejandro Ceron-Noriega
- Supervisor: René Ketting
- Co-Supervisors: Susanne Gerber
- Research Group
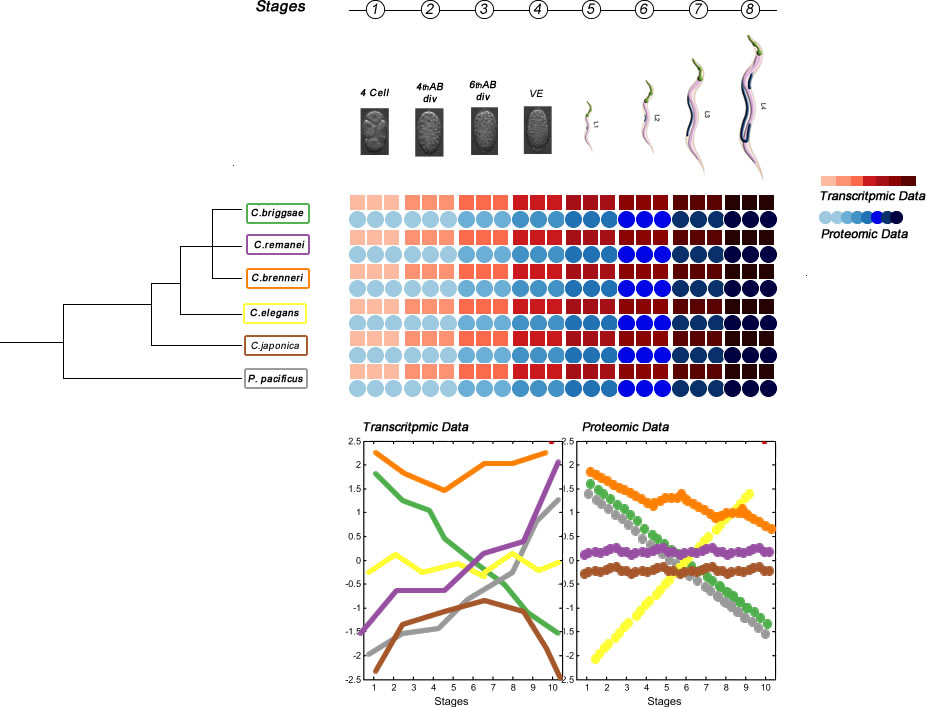
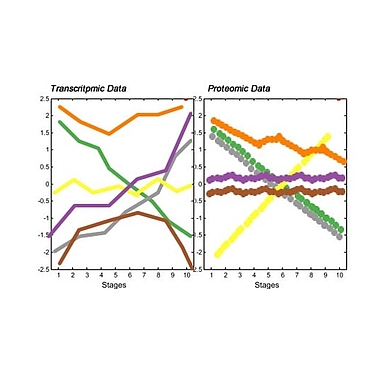
My PhD project focuses on developmental dynamics of transcription and protein expression in embryos of six species of the genus Caenorhabditis, in particular on evolutionary changes across these different species. To achieve this goal, we need a reliable and high-quality protein-coding gene annotation.
My PhD project focuses on developmental dynamics of transcription and protein expression in embryos of six species of the genus Caenorhabditis, in particular on evolutionary changes across these different species. However, the developmental time courses of transcription dynamics have already been raised by a large variety of research. These include studies in C. elegans (Baugh 2003), Drosophila melanogaster (Brown 2014), the comparison among these two and zebrafish (Mathavan 2005). A time course including the same species as my project was executed by (Levin 2012). Focusing only on transcriptome dynamics, though revealing interesting features, has clear limitations. Transcript levels are only moderate predictors for protein expression (Prasad 2017). The principal weakness is that the approach does not take post-transcriptional processes such as translational regulation or protein stability into account. The main contribution of my project is the parallel production of transcriptome and proteome data, which will be directly comparable.To achieve this goal, we need a reliable and high-quality protein-coding gene annotation. Since some of the target species do not have a proper annotation available, the first effort of my PhD project aims to improve the annotation of these species. Considering that this effort involves the establishment of a bioinformatic pipeline, access to RNA-sequencing and proteomics facilities and since some of the target species lack accurate annotation in Wormbase version WS273, the first effort of my PhD project will combine in-depth whole transcriptome sequencing and high-resolution mass spectrometry to improve gene annotation of protein-coding genes in six Caenorhabditis species: Caenorhabditis elegans and its sister species Caenorhabditis inopinata (Woodruff et al. 2017), Caenorhabditis brenneri, Caenorhabditis remanei and Caenorhabditis briggsae; Caenorhabditis japonica and 7 species that are phylogenetically interesting which lack reference genomes in the current Wormbase database.