Project 23.3
- PhD student: Carl Weile
- Supervisor: Jan Padeken
- Co-Supervisors: Katharina Papsdorf
- Further TAC-members: Meret Huber, René Ketting
- Research Group
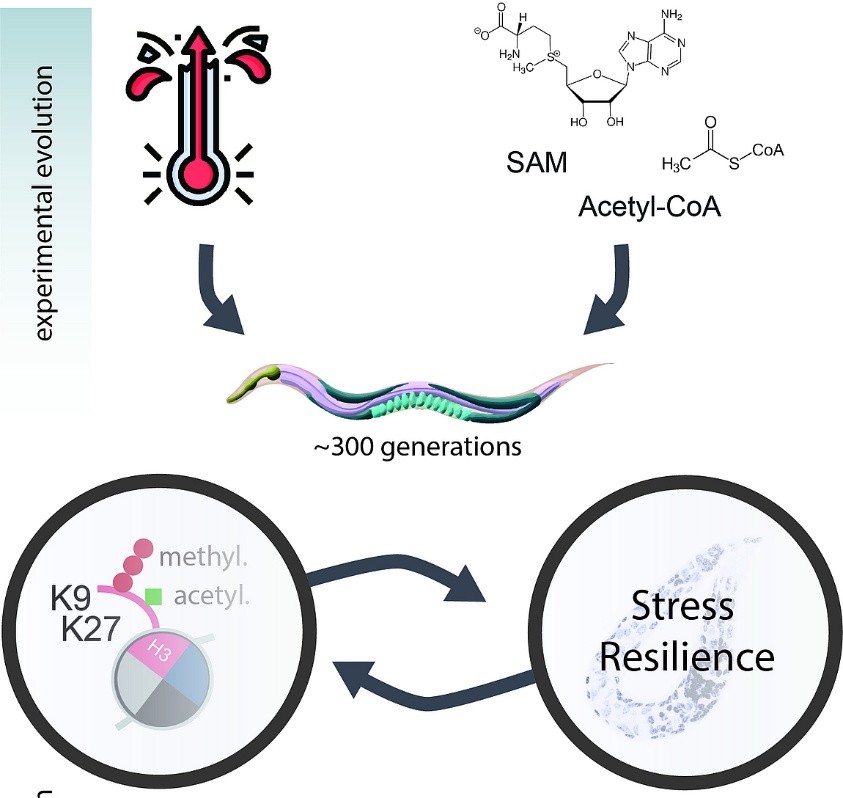
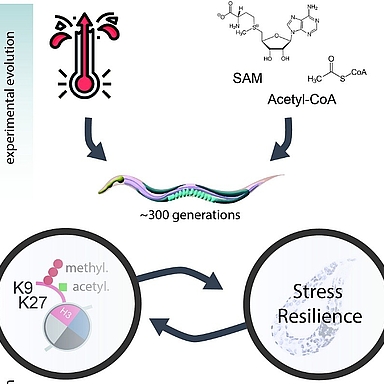
Organisms adapt to environmental stress through genetic mutations and epigenetic changes. Epigenetic modifications can influence traits across generations. By running a multigenerational experiment, in C. elegans, that tracks phenotypic responses to stress alongside genetic and epigenetic changes, we can explore how epigenetics contributes to adaptation, whether it supports genetic change, and if these modifications happen randomly or are directed. This helps us better understand the role of epigenetics in long-term adaptation.
In nature, organisms adapt to environmental stressors to survive and thrive. While genetic adaptation through random mutations is well understood, epigenetic changes can also, in principle, drive adaptation to stress (Jaenisch & Bird, 2003; Heard & Martienssen, 2014). However, many open questions remain regarding epigenetic adaptation, such as:
• How do epigenetic changes contribute to adaptation?
• Do they support or trigger genetic adaptation?
• When do they occur in relation to DNA mutations?
• Where in the genome do they arise?
• Are these changes directed by the environment, or are they random?
To address these questions, we are conducting a multigenerational experiment tracing phenotypic responses to environmental stress and linking them to underlying genetic and epigenetic changes. This allows us to map the molecular basis of adaptation over time and investigate where, when, and how adaptation occurs on both genetic and epigenetic levels (Rechavi & Lev, 2017; Cavalli & Heard, 2019).
For this purpose, we are using the model organism Caenorhabditis elegans (C. elegans). Its short generation time of 3–4 days makes it ideal for studying adaptation across multiple generations in a relatively short timeframe (Brenner, 1974). By transferring approximately 10,000 larvae each generation, we ensure clear generational separation and minimize bottleneck effects, maintaining population diversity and statistical robustness (Frézal & Félix, 2015).
To simulate environmental stress, we are cultivating C. elegans under different conditions. One condition involves raising them at 25°C, which constitutes continuous heat stress compared to their standard cultivation temperature of 20°C (Glauser et al., 2011). Another stressor is a change in diet: instead of the typical E. coli OP50 strain, we are using the HT115 strain. HT115 is lower in nutrients, placing the worms under nutritional stress (Vrablik & Watts, 2013).
To track phenotypic adaptation, we will perform stress assays in every generation. These assays measure the population’s sensitivity to specific stressors, allowing us to determine if and when adaptation occurs by comparing performance over generations (Rodriguez et al., 2013).
In parallel, we will conduct molecular analyses to uncover the epigenetic and genetic basis of any observed phenotypic changes. We currently plan to use RNA-seq, small RNA-seq (sRNA), ATAC-seq, and CUT&Tag to investigate transcriptional changes, chromatin accessibility, and histone modifications (Buenrostro et al., 2013; Kaya-Okur et al., 2019).
This integrative approach will help us understand how epigenetic mechanisms contribute to adaptation, whether they play a role in facilitating genetic change, and to what extent they are random or environmentally guided. Ultimately, this project aims to shed light on the dynamic relationship between phenotype, epigenetics, and the environment across generations.