Project 9.2
- PhD student: Vanessa Maißl
- Supervisor: Helen May-Simera
- Co-Supervisors: Susanne Foitzik, Susanne Gerber
- Research Group
This project focuses on the non-ciliary functions of ciliary proteins. Primary cilia are organelles that protrude from the surface of most cell types. Proteins localised to the primary cilium often have functions predominantly related to the cilium. However, it has been shown that ciliary proteins can also have non-ciliary functions. The localisation inside of the nucleus is particularly interesting. The functions of these proteins here are still largely unexplored, which is why this project addresses the question which role these proteins play inside the nucleus.
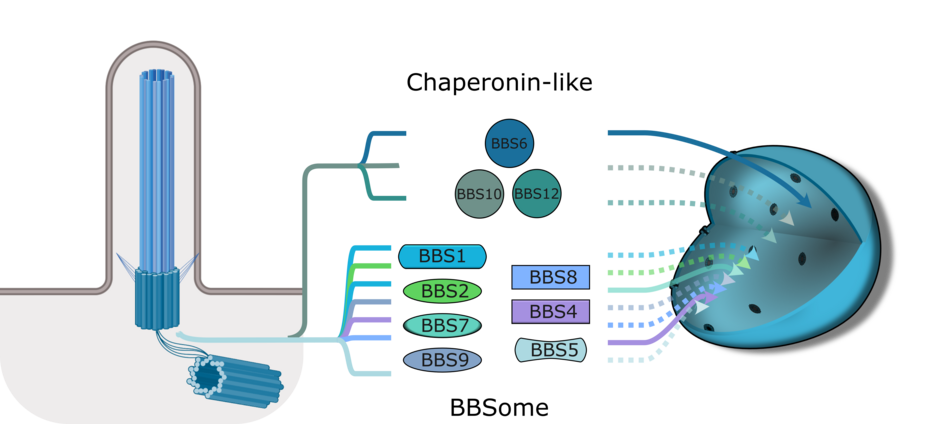
Primary cilia play an important role in cell signalling and development. Mutations affecting the primary cilium can disrupt tissue or organ homeostasis and lead to diseases known as ciliopathies. One of these ciliopathies is Bardet-Biedl syndrome (BBS), which is considered an archetypal ciliopathy because patients have all the phenotypes associated with dysfunction of the primary cilia. Mutations have been identified in over 23 proteins associated with BBS, many of which form two main complexes: the BBSome and the chaperonin-like complex. BBS1, BBS2, BBS4, BBS5, BBS7, BBS8, BBS9 and BBS18 belong to the BBSome trafficking complex, while BBS6, BBS10 and BBS12 belong to the chaperonin-like complex that helps assemble the BBSome.
In this project, we aim to answer two main questions: first, whether stress-induced nuclear import is also a feature of BBS proteins outside animals, and second, what are the consequences of re-localisation of BBS proteins to the nucleus and how does it affect gene regulatory functions in animal and non-animal cells. To answer the first question we will label BBS proteins in cells of eukaryotes (Crithidia) and investigate their nuclear localisation after different treatments by immunocytochemistry and/or Western blot.
For the second question, we will further investigate the role of BBS in gene regulation. We have already identified numerous interactions between BBS proteins and DNA regulators. We will confirm the interaction of DNMT1 with one of the BBS proteins, to investigate the potential role of BBS in methylation. In a transcriptomic approach, we will overexpress BBS proteins in the nucleus as well as knock out BBS and look for changes in gene regulation and expression.
These two approaches will be performed in both mammalian (human, mouse) and non-mammalian (Crithidia) eukaryote cell culture models.