Project 5.3
- PhD student: Mohd Aaquib
- Supervisor: René Ketting
- Co-Supervisors: Hanna Kokko
- Further TAC-members: Meret Huber
- Research Group
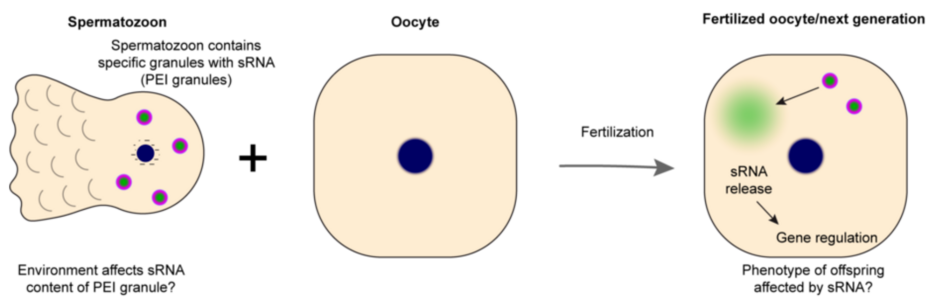
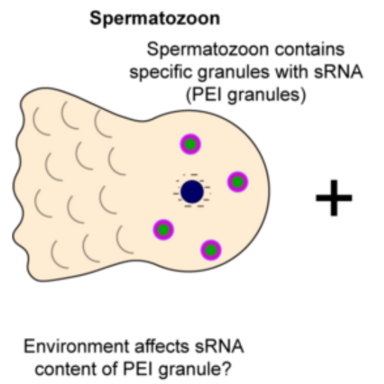
Small RNAs play crucial role in developmental gene regulation and genome protection against harmful genetic elements. In C. elegans and many other organisms, they can mediate transgenerational gene silencing. Since the transmission of gene regulatory states is vital for adapting to environmental challenges, we aim to explore whether C. elegans can use small RNA-based transgenerational inheritance as a mechanism to adapt to changing environmental cues, such as variations in nutrition and temperature.
In animals, small RNAs work in concert with Argonaute proteins, functioning as a guide and an effector respectively. They typically dampen the gene expression by targeting mRNA, either through endonucleolytic cleavage or other transcriptional or translational control. RNA interference proceeds in two steps in C. elegans, first small RNAs undergo target-dependent amplification generating so called secondary small RNAs which mediate the gene silencing. These secondary small RNAs (known as 22G RNAs) associate with Argonautes (like WAGO-3) that enable their transmission via germ cells to the progeny. The distinct steps take place within confined spaces called germ granules which are biomolecular condensates.
The project investigates how small RNA-mediated gene regulation in C. elegans responds to environmental changes and whether this response can be inherited. While RNA interference (RNAi) and small RNA inheritance are well established in this model, the exact molecular underpinnings-especially regarding their role in evolutionary adaptation remain unclear.
The central idea is that environmental conditions, such as alteration in diet or temperature may shape the small RNA landscape and affect gene regulatory states. Small RNAs in association with Argonautes might then be transmitted to the progenies influencing gene regulation in future generations that help in adapting to the environment.
We plan to utilise C. elegans, a fast-developing model organism with excellent genetic tool, and expose to varying environmental conditions-specifically different bacterial strains and temperatures and collect RNA population across several generations. We plan to utilise small RNA-seq, and RNA-immunoprecipitation sequencing (RIP-seq) to identify alterations in small RNA profile and inheritance-related Argonaute associated RNA population. We will further utilise CRISPR based genetic approaches and Super resolution confocal Microscopy to address molecular mechanisms of possible transgenerational inheritance.