Project 5.1
- PhD student: Shamitha Govind
- Supervisor: René Ketting
- Co-Supervisors: Hans Zischler
- Further TAC-members: Joan Barau , Peter Sarkies
- Research Group
Evolutionary arms race with transposable elements has greatly diversified the sRNA pathways in individual species. This resonates in the GTSF-1 proteins, which are essential for sRNA pathways in many species but show evolutionary plasticity by acting at different steps of the pathway in different species. In this project, we are characterizing GTSF-1 from three nematodes namely C. inopinata, C. briggsae and P. pacificus using transgenic techniques, transcriptomics and proteomics. We plan to derive a general molecular function of GTSF-1 and gain insight into the evolution of molecular pathways in sRNA biology.
Small-RNAs (sRNAs) are important regulators of gene expression. By binding to an Argonaute (Ago) protein, the sRNA-Ago complex can interfere with mRNA function, mostly through transcriptional or translational silencing. sRNA pathways are found in all clades of life and various classes of regulatory sRNAs have been described. While these share a common function in regulating transposable elements, the pathways themselves are evolutionarily labile and their molecular organization vary substantially between different species. Even well conserved proteins can take up different roles depending on the species.
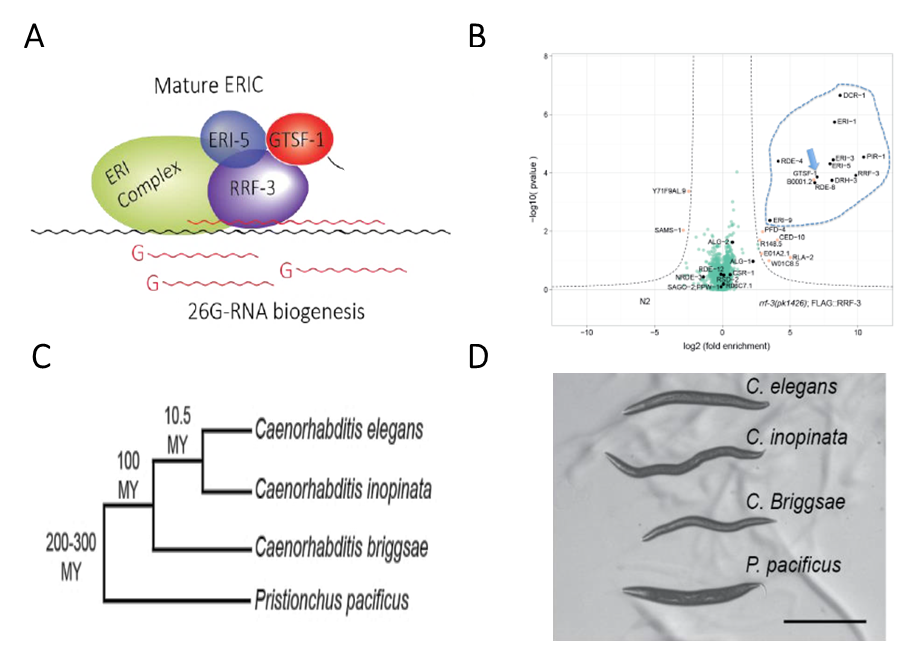
A well-studied example of this flexibility is GTSF-1, a highly conserved 20kDa long Zinc finger protein. In Drosophila melanogaster and Mus musculus, GTSF-1 participates in a transposon-silencing sRNA pathway. In D. melanogaster, GTSF-1 is a downstream factor where it binds a nuclear Ago and drive transcriptional silencing of transposon [1]. However, in M. musculus, GTSF-1 acts upstream of the pathway by binding a cytoplasmic Ago and enabling the biogenesis of sRNAs [2]. Surprisingly, in C. elegans, GTSF-1 is not involved in transposon silencing and does not bind Ago proteins. It instead forms a protein complex with an RNA Dependent RNA polymerase (RdRP) called RRF-3 [3]. Together they facilitate the biogenesis of another class of small RNAs, which ultimately targets pseudogenes and other recent gene duplications.
This striking functional plasticity of GTSF-1 led us to hypothesize that even closely-related species have very customized sRNA populations. We are curious whether biochemical building blocks that together make a sRNA pathway can be differentially used to better meet the specific needs for genome defense in individual species.
To address this, we are studying sRNA pathways in a set of relatively closely related nematodes C. inopinata, C. briggsae and P. pacificus. The genomes of these nematodes have been sequenced and homologous RNAi factors and sRNA populations have been profiled [4]. They have similar RNAi-like pathways to C. elegans but specific individual targets and sRNA sequences are not conserved. GTSF-1 is one of the well-conserved sRNA pathway factors and the mentioned nematodes each carry a single homologous gtsf-1 gene. In this project, we wish to characterize the molecular function of GTSF-1 proteins in these nematodes using transgenic techniques, transcriptomics and proteomics.
We are using CRISPR/Cas-9 and RNA interference to modify GTSF-1 and other sRNA pathway components in these nematodes while also developing specific antibodies against the homologous GTSF-1 proteins. This will allow us to compare differences in growth, fertility and sRNA populations of mutants to their C. elegans counterpart and shed light on the role of GTSF-1 in these species. We are using the antibodies to perform Immunoprecipitation coupled to mass-spectrometry to test whether the interaction of GTSF-1 –RRF-3 is unique to C. elegans or a broadly observed feature in nematodes. This is particularly interesting because RRF-3-like RdRP orthologues are distributed throughout the nematode phylum, suggesting that ancestral nematode sRNA biogenesis was perhaps similar to RRF-3-mediated sRNA biogenesis in C. elegans. Finally, using CRISPR/Cas-9, we are making cross-species complement strains to test activity of C. briggsae, C. inopinata and P. Pacificus- GTSF-1 in C. elegans and study the extent of functional conservation of GTSF-1 proteins.
The combined studies will describe a general molecular function of GTSF-1 like proteins in small RNA biology. Further, it will reveal how small RNA pathways have evolved during nematode evolution in relation to genome characteristics and gene-regulatory needs of the individual species.
References:
1. Dönertas, D., Sienski, G. & Brennecke, J. Drosophila Gtsf1 is an essential component of the Piwi-mediated transcriptional silencing complex. Genes Dev. 27, 1693–1705 (2013).
2. Ohtani, H. et al. DmGTSF1 is necessary for Piwi-piRISC-mediated transcriptional transposon silencing in the Drosophila ovary. Genes Dev. 27, 1656–1661 (2013).
3. Yoshimura, T. et al. Mouse GTSF 1 is an essential factor for secondary piRNA biogenesis. 1–17 (2018) doi:10.15252/embr.201642054.
4. Almeida, M. V. et al. GTSF‐1 is required for formation of a functional RNA‐dependent RNA Polymerase complex in Caenorhabditis elegans. EMBO J. 37, e99325 (2018).
5. Sarkies, P. et al. Ancient and Novel Small RNA Pathways Compensate for the Loss of piRNAs in Multiple Independent Nematode Lineages. PLoS Biol. 13, 1–20 (2015).