Padeken/Papsdorf
Adaptation to environmental perturbations using natural C. elegans isolates
Supervisor: Jan Padeken
Co-Supervisor: Katharina Papsdorf
Scientific Background
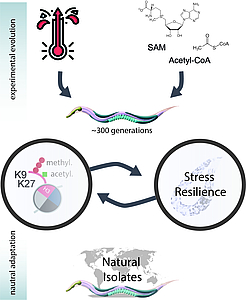
How do environmental stresses shape the epigenome? Nematode worms and C. elegans in particular are well established model organisms for molecular biology, extensively studied under laboratory conditions. In the wild, however, C. elegans have been isolated from very diverse habitats that expose them to several -sometimes –extreme- environmental perturbations. These include different diets, temperatures, and UV radiation. For example, C. elegans has been isolated from habitats with drastically different temperatures such as Kenya and Switzerland, or food sources like nuts and shellfish. Classically adaptation to these perturbations has been studied in the context of DNA mutations. Gene expression however can also be adapted by changes in the chromatin signature. Importantly chromatin marks are directly connected with nutrient availability via shared cofactors from metabolic pathways. For example, acetyl-CoA, which is a degradation product of fatty acid oxidation is used by histone acetyltransferases as a cofactor to add acetyl groups to lysine residues on histones. In addition, histone methyl transferases use the metabolite S-Adenosyl methionine (SAM) as a methyl donor. This allows the organism to directly integrate signals from nutrients into their gene expression. In the case of temperature stress, we recently showed that the enzymes catalyzing heterochromatin transiently disperse during heat stress, resulting in a widespread activation of genes, essential for the resistance to heat exposure. However, whether and how this interplay between nutrient availability and temperature stress with epigenetic changes contributes to the long-term adaptation of an organism is unclear.
Project description
Our project addresses two primary questions. First, we will exploit the rapid generation time of laboratory cultured C. elegans wild-type and chromatin mutant strains under conditions of nutritious stress (low and high fat diet) and
temperature stress (low and high). At regular intervals, we will perform established assays to assess the resilience of these continuously ongoing cultures to increased stress levels. We will focus on starvation-resistance, acute heat-shock response, lifespan, and organismal fitness, including developmental timing and brood size. Concurrently, we will collect samples to analyze chromatin modifications (using CUT&Run and ATAC-seq), transcriptional changes (via RNA-seq), and metabolite levels with a direct link to histone modifications (including lipids, Acetyl-CoA, and SAM). This approach allows us to track the molecular adaptation processes over time and correlate the changes with phenotypic variations and overall fitness.
In a complementary strategy, we will utilize the extensive collection of natural C. elegans isolates maintained by the research community (https://caendr.org/). These nematodes have been collected from diverse climatic regions worldwide. We have selected natural isolates from habitats with a wide range of ambient temperatures and food sources, such as Kenya, southern Spain, the coasts of Australia, Switzerland, Peru, and Washington state (USA). By profiling chromatin marks, organismal fitness, and stress responses in these isolates, analogous to our first approach, we aim to determine which aspects of laboratory-controlled adaptation are reflected in natural environments.
Together, these two approaches will elucidate the role of the epigenome in an organism's adaptation to environmental stresses and provide insights into the interplay between metabolic and epigenetic homeostasis.
We are looking for a highly motivated team player, interested in combining genomics with evolutionary aspects in a project that has the potential to address a fundamental question in the epigenetics field and bring a new aspect to evolutionary biology.
What you will learn
Conceptual
• Understanding the connections between gene regulation, stress response and organismal fitness
• Epigenetic and Genetic adaptation
• Heterochromatin biology and epigenetics.
Practical skills
• Cutting edge epigenome profiling techniques (CUT&Tag, ATAC-seq).
• Profiling key metabolites.
• Integration of multiple genomics datatypes with quantitative phenotyping and assays for stress resilience
Your qualifications
We are looking for a highly motivated team player, interested in combining genomics with evolutionary aspects. You will have a chance to handle and combine multiple datatypes, such as CUT&Run, Whole genome sequencing, RNA-seq, lifespan assays, stress response, developmental timing, fertility assays.
Publications relevant to this project
Delaney CE, Methot SP, Kalck V, Seebacher J, Hess D, Gasser SM, Padeken J (2022) SETDB1-like MET-2 promotes transcriptional silencing and development independently of its H3K9me-associated catalytic activity. Nat Struct Mol Biol, 29:85–96. https://doi.org/10.1038/s41594-021-00712-4
Delaney CE, Methot SP, Guidi M, Katic I, Gasser SM, Padeken J (2019) Heterochromatic foci and transcriptional repression by an unstructured MET-2/SETDB1 co-factor LIN-65. J Cell Biol. Mar 4;218(3):820-838. https://doi.org/10.1083/jcb.201811038
Papsdorf K, Miklas JW, Hosseini A, Cabruja M, Morrow CS, Savini M, Yu Y, Silva-García CG, Haseley NR, Murphy LM, Yao P, de Launoit E, Dixon SJ, Snyder MP, Wang MC, Mair WB, Brunet A (2023) Lipid droplets and peroxisomes are co-regulated to drive lifespan extension in response to mono-unsaturated fatty acids. Nat Cell Biol, 25:672-684. https://doi.org/10.1038/s41556-023-01136-6
Papsdorf K and Brunet A (2019) Linking lipid metabolism to chromatin regulation in aging. Trends Cell Biol, 29:97-116. https://doi.org/10.1016/j.tcb.2018.09.004
Singh PP, Reeves GA, Contrepois K, Papsdorf K, Miklas JW, Ellenberger M, Hu CK, Snyder MP, Brunet A (2024) Evolution of diapause in the African turquoise killifish by remodeling the ancient gene regulatory landscape. Cell. 187(13):3338-3356.e30. https://doi.org/10.1016/j.cell.2024.04.048