May-Simera/Colgan/Barau/Foitzik
Co-option of cilia proteins in gene regulation
Supervisor: Helen May-Simera
Co-Supervisor: Joseph Colgan, Joan Barau, Susanne Foitzik
Scientific Background
Cilia and flagella are widely present across eukaryotic lineages and evolved to serve a multitude of functions, from locomotion and chemotaxis in unicellular green algae to reception of light in animals. Although their functions vary, many proteins required to build and maintain these structures are conserved across species. More recently, ciliary proteins have been found to localise to other cellular compartments where they may function in non-ciliary processes. These have most likely arisen via a process of co-option. One of these non-ciliary processes is likely to include gene regulation, since a subset of ciliary proteins have been shown to interact with DNMT1 and modulate DNA Methylation.
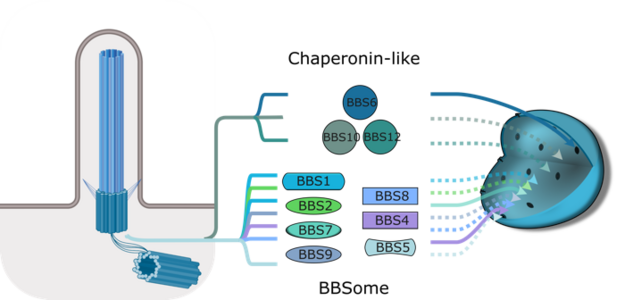
In our lab we focus on the Bardet-Biedl Syndrome (BBS) proteins, a special class of ciliary trafficking molecules. Genetic mutations in BBS genes leads to a multitude of pathogenic symptoms in patients. We previously identified orthologous BBS proteins across a diverse phylogenetic spectrum and bioinformatically predicted the presence of putative nuclear localization or nuclear exit signals (NES/NLS). Importantly, these seem to have evolved independently of the mode of mitosis, which may suggest an evolutionary selected active process. We were also able to show that BBS proteins have the capacity to enter the nucleus particularly upon stress situations in animal cells.
Project description
With this PhD Project we hope to address two distinct questions:
Q1) Is stress-related nuclear import also a feature of BBS proteins outside of animals?
For this we will expand our previous exanimations looking for the presence of BBS proteins in the nucleus in non-model organisms (Chrithidia, Chlamydomonas, Ants, Bees), via both bioinformatic as well as wet laboratory approaches. The choice of proteins and organisms to analyse will be based on bioinformatic predictions and technical capabilities.
Q2) What is the consequence of BBS protein re-localization to the nucleus and how do they influence gene regulatory functions?
To address Q2 we will adopt two main approaches. First, a proteomic approach in which we will identify the BBS nuclear interactome via co-immunoprecipitation of tagged BBS proteins from nuclear fractions and subsequent Mass Spectrometry. Second a transcriptomic approach in which we will overexpress BBS proteins in the nucleus and look for changes in gene regulation and expression.
Since we have already seen that stress can influence the nuclear localization of these proteins, we will repeat these experiments upon induction of various cellular stresses.
What you will learn
Standard Molecular Biology techniques, Phenotyping of various disease models (including patient derived model systems)
Your qualifications
Masters in Biology or Molecular Biology. Motivation and Creativity highly valued.
Publications relevant to this project
Ewerling A, Maissl V, Wickstead B, May-Simera HL (2023). Neofunctionalization of ciliary BBS proteins to nuclear roles is likely a frequent innovation across eukaryotes. iScience. 26(4):106410. doi.org/10.1016%2Fj.isci.2023.106410
Brücker L, Becker SK, Maissl V, Harms G, Parsons M, May-Simera HL (2023) The actin-bundling protein Fascin-1 modulates ciliary signalling. J Mol Cell Biol. 15(4):mjad022. doi.org/10.1093/jmcb/mjad022
Patnaik SR, Farag A, Brücker L, Volz AK, Schneider S, Kretschmer V, May-Simera HL (2020). Tissue-dependent differences in Bardet-Biedl syndrome gene expression. Biol Cell. 112(2):39-52. doi.org/10.1111/boc.201900077