Ketting/Kokko
Inheritance of epigenetic information via small RNA molecules
Supervisor: René Ketting
Co-Supervisor: Hanna Kokko
Scientific Background
Gene regulation by small non-coding RNAs is a deeply conserved mode of gene control. It is used in developmental control of gene expression, but also in the protection of the genome against mobile genetic elements (i.e. transposons). The small RNA molecules function as sequence specific guides for proteins of the Argonaute family, which can repress translation, induce endonucleolytic cleavage or induce heterochromatin upon recognition of transcripts with sequence homology to the small RNA. Hence, the small RNA co-factors are essential players in these pathways as they fully determine the specificity. We are studying various aspects of small RNA-driven gene regulation using two model organisms: the nematode C. elegans and the zebrafish.
In the context of GenEvo we are interested in how small RNA molecules may be used to transfer gene regulatory information (epigenetic information) from one generation to the next. In particular, we want to address how and if environmental factors such as nutrition or temperature can be translated into pools of small RNA molecules that help to adapt to the circumstances at hand, and whether these can then be transmitted to offspring.
Project description
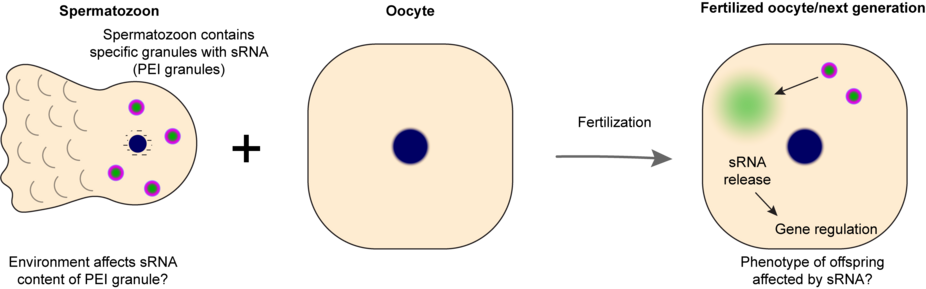
During meiosis, most regulatory marks on the chromatin are erased and newly established in the next generation. If environmental influence is to have an effect on gene regulation in a next generation, somehow information needs to be transferred between parent and offspring. Small RNAs are strong candidates for such activities, and in some cases such inheritance indeed has been described. However, how environmental factors can translate into small RNA populations in the germ cells, that can then be transmitted into the offspring is largely unknown. Likewise, to what extent such inheritance can be expected to affect evolutionary processes is poorly understood.
Based on previously published work describing sperm-specific condensates that act in paternal inheritance, we study how small RNAs can be inherited through the male germline. Specifically, we study this using the nematode C. elegans. By understanding the molecular principles of this process, it can possibly be manipulated in a precise manner, without touching other aspects of small RNA biology. Thus, we aim to establish a system in which we will be able to separate the effect of inheritance of small RNAs from many potential secondary effects, and to be able to study how/whether small RNA inheritance has the potential to contribute to evolutionary adaptation to environmental stresses.
In this project, you will be studying how small RNAs can be specifically loaded into sperm, what their impact can be in zygotes and embryos, and whether environmental cues may be inherited in this manner in order to help adapt the offspring to the circumstances that were experienced by their parents. In collaboration with the group of prof. Kokko we will test under which regimes such inheritance can be relevant for adaptation to environmental conditions. We will use experiments to test models and to provide data that can feed back into the models.
What you will learn
Techniques that will be used in this project are:
- microscopy
- genetics
- CRISPR-Cas genome editing
- RNA interference
- RNAseq
- analysis of sequencing data
- theory development (Kokko collaboration)
Your qualifications
We are looking for people with a strong natural curiosity, perseverance and a high intrinsic motivation. Specific knowledge of the applied techniques is not essential, but a strong basis in molecular biology is required. We offer a highly interactive, stimulating and flexible research environment, with ample opportunities for personal development.
Publications relevant to this project
Podvalnaya N*, Bronkhorst AW*, Lichtenberger R, Hellmann S, Nischwitz E, Falk T, Karaulanov E, Butter F, Falk S#, Ketting RF# (2023) piRNA processing by a trimeric Schlafen-domain nuclease.Nature 622(7982):402-409. doi.org/10.1038/s41586-023-06588-2
Schreier J, Dietz S, Boermel M, Oorschot V, Seistrup AS, de Jesus Domingues AM, Bronkhorst AW, Nguyen DAH, Phillis S, Gleason EJ, L’Hernault SW, Phillips CM, Butter F, Ketting RF (2022) Membrane-associated cytoplasmic granules carrying the Argonaute protein WAGO-3 enable paternal epigenetic inheritance in Caenorhabditis elegans.Nat. Cell Biol. 24(2):217-229. doi.org/10.1038/s41556-021-00827-2
Marnik EA*, Almeida MV*, Cipriani PG*, Chung G, Caspani E, Karaulanov E, Gan HH, Zinno J, Isolehto IJ, Kielisch F, Butter F, Sharp CS, Flanagan RM, Bonnet FX, Piano F, Ketting RF#, Gunsalus KC# and Updike DL# (2022) The Caenorhabditis elegans TDRD5/7-like protein, LOTR-1, interacts with the helicase ZNFX-1 to balance epigenetic signals in the germline.PLoS Genet. 18:e1010245. doi.org/10.1371/journal.pgen.1010245
Schreier J, Ketting RF (2022) How stress can affect your sex appeal. (Preview)Dev. Cell 57(3):291-292. doi.org/10.1016/j.devcel.2022.01.009
Ketting RF, Cochella L (2021) Concepts and functions of small RNA pathways in C. elegans. (Review)Curr. Top. Dev. Biol. 144:45-89. doi.org/10.1016/bs.ctdb.2020.08.002