Ketting/Barau/Burger
TFs to TEs: the emergence of novel regulatory networks from the antagonistic interaction between transcription factors and transposable elements
Supervisor: René Ketting
Co-Supervisor: Joan Barau, Joachim Burger
Scientific Background:
A body of evidence suggests that Transposable Elements (TEs) can act as key contributors to the emergence of novel gene expression patterns1. TEs harbor several potential transcription factor (TF) binding sites in their sequences and are targets of a plethora of epigenetic regulatory mechanisms and reader proteins2. TE copy number and mobility can leverage these characteristics into the creation of entirely new gene regulatory networks with long-term consequences to the evolution of genome regulation in the context of development and homeostasis1,2.
PhD project: TFs to TEs: the emergence of novel regulatory networks from the antagonistic interaction between transcription factors and transposable elements
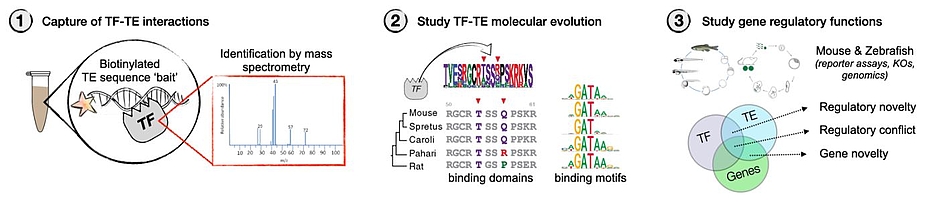
We propose to generate a framework to understand how TE-derived contributions to the evolution of gene regulation can evolve from the parasitic relationship TEs have with genomes. We will use mass spectrometry to identify the TFs potentially binding to TE bait sequences in germ cell extracts from vertebrate species, the archetypal context where selfish, antagonistic TF-TE interactions occur. We will then look for signatures of positive selection in TE promoter sequences and cognate TF DNA binding domains, focusing on validating the changes in motif preference and how this impacts the range of DNA targets. We will finally expand to explore predicted targets with potential regulatory function outside of the genetic conflict context of germ cells, and attempt to characterize stabilizing or antagonizing epigenetic mechanisms that may enable these as novel genome regulatory mechanisms. Previously using similar approaches, we have identified NAIF13, a TF that evolved from a coopted Harbinger transposon, is conserved in vertebrates and whose DNA binding is dependent on epigenetic modifications of its cognate binding motif. We will use NAIF1 as a key example of TE-sequence binding protein and use a combination of mouse and zebrafish molecular and developmental biology to uncover its roles in development. Project execution will involve 3 major steps (Figure1):
1. Vertebrate-focused discovery of TE promoter binding proteins in collaboration with Dr. Falk Butter. Select and validate candidate-species interactions by ChIP or CUT&RUN in target species. This part of the project will involve efforts from both the Barau and Ketting labs related to material collection, processing, and analysis.
2. Study the patterns of molecular evolution of the DNA binding domains and their cognate motifs in silico and in vitro. Select and validate candidates in vitro or in tissue culture experiments when possible. This part of the project will take place in the Barau lab and involve a collaboration with the IMB protein expression core facility.
3. Study selected candidates for potential function of the uncovered interactions in gene regulation and development (focus on mouse & zebrafish). This part of the project will involve both the Barau (mouse) and Ketting (Zebrafish) labs. As an outlook, we would like to use the knowledge about the experimentally observed patterns of co-evolution between TFs and motifs at TEs to computationally mine modern and ancient available hominid genomes for similar signatures of evolution across predicted TF binding sites at TEs and the coding sequences of their cognate DNA binding domains encoded in the TF genes. This part will benefit from co-supervision of Prof. Dr. Joachim Burger.
Publications relevant to this project
1. Feschotte. 2008. “Transposable Elements and the Evolution of Regulatory Networks.” Nat Rev Genet 9 (5): 397–405.
2. Hermant & Torres-Padilla. 2021. “TFs for TEs: The Transcription Factor Repertoire of Mammalian Transposable Elements.” Genes & Dev 35 (1-2): 22–39.
3. Sinzelle et al., 2008. “Transposition of a Reconstructed Harbinger Element in Human Cells and Functional Homology with Two Transposon-Derived Cellular Genes.” PNAS 105 (12): 4715–20.